Jupyter Notebook Best Practices for Data Science
September 10th, 2015
The Jupyter Notebook is a fantastic tool that can be used in many different ways. Because of its flexibility, working with the Notebook on data science problems in a team setting can be challenging. We present here some best-practices that SVDS has implemented after working with the Notebook in teams and with our clients.
The need to keep work under version control, and to maintain shared space without getting in each other’s way, has been a tricky one to meet. We present here our current view into a system that works for us—and that might help your data science teams as well.
Overall thought process
There are two kinds of notebooks to store in a data science project: the lab notebook and the deliverable notebook. First, there is the organizational approach to each notebook.
Lab Notebooks:
Let a traditional paper lab notebook be your guide here:
- Each notebook keeps a historical (and dated) record of the analysis as it’s being explored.
- The notebook is not meant to be anything other than a place for experimentation and development.
- Each notebook is controlled by a single author: a data scientist on the team (marked with initials).
- Notebooks can be split when they get too long.
- Notebooks can be split by topic, if it makes sense.
Deliverable Notebooks:
Deliverable notebooks are the fully polished version of the lab notebooks, and store the final outputs of analysis. These notebooks are controlled by the whole data science team, rather than by any one individual.
Version Control
Here’s an example of how we use git and GitHub. One beautiful new feature of Github is that they now render Jupyter Notebooks automatically in repositories.
When we do our analysis, we do internal reviews of our code and our data science output. We do this with a traditional pull-request approach. When issuing pull-requests, however, looking at the differences between updated .ipynb files, the updates are not rendered in a helpful way. One solution people tend to use is to commit the conversion to .py instead. This is great for seeing the differences in the input code (while jettisoning the output), and is useful for seeing the changes. However, when reviewing data science work, it is also incredibly important to see the output itself.
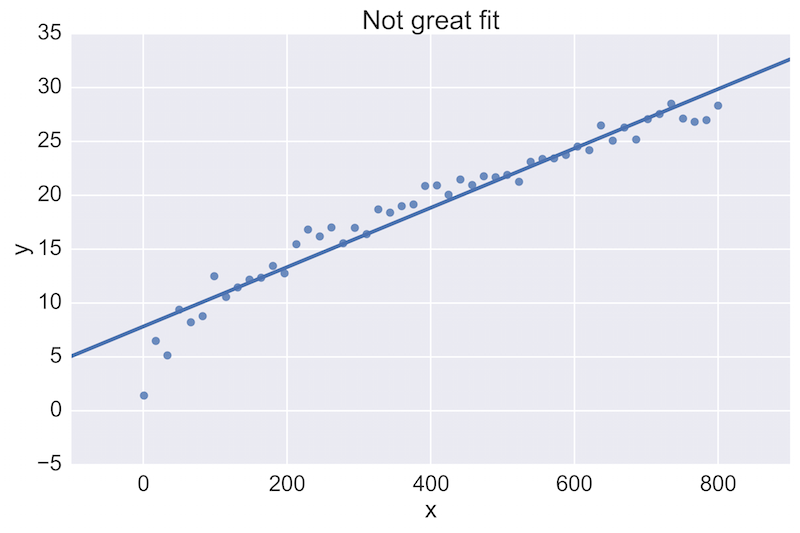
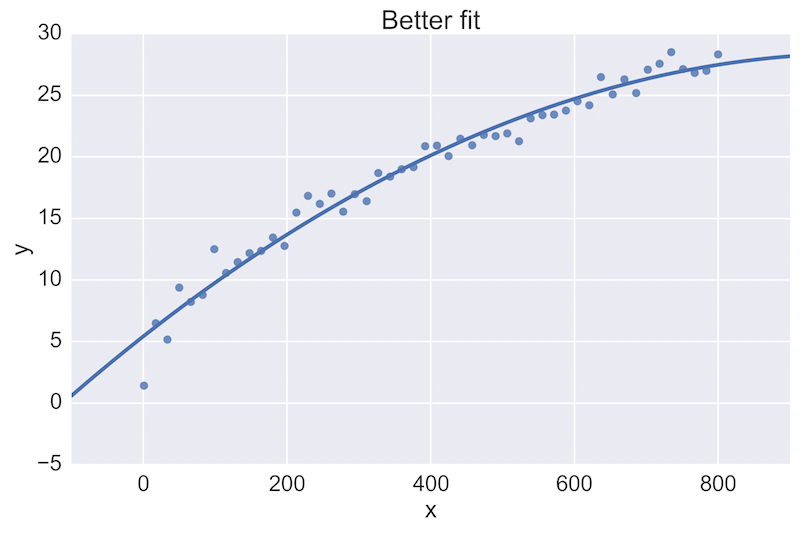
For example, a fellow data scientist might provide feedback on the following initial plot, and hope to see an improvement:
The plot on the top is a rather poor fit to the data, while the plot on the bottom is better. Being able to see these plots directly in a pull-request review of a team-member’s work is vital.
See the Github commit example here.
Take the time to notice that there are three ways to see the updated figure.
Post-Save Hooks:
We get around these difficulties by committing the .ipynb, .py, and .html of every notebook. Creating the .py and .html files can be done simply and painlessly by editing the config file:
~/.ipython/profile_nbserver/ipython_notebook_config.py
and adding the following code:
### If you want to auto-save .html and .py versions of your notebook:
# modified from: https://github.com/ipython/ipython/issues/8009
import os
from subprocess import check_call
def post_save(model, os_path, contents_manager):
"""post-save hook for converting notebooks to .py scripts"""
if model['type'] != 'notebook':
return # only do this for notebooks
d, fname = os.path.split(os_path)
check_call(['ipython', 'nbconvert', '--to', 'script', fname], cwd=d)
check_call(['ipython', 'nbconvert', '--to', 'html', fname], cwd=d)
c.FileContentsManager.post_save_hook = post_save
Now every save to a notebook updates identically-named .py and .html files. Add these in your commits and pull-requests, and you will gain the benefits from each of these file formats.
Putting it all together
Here’s the directory structure of a project in progress, with some explicit rules about naming the files.
Example Directory structure:
- develop # (Lab-notebook style)
+ [ISO 8601 date]-[DS-initials]-[2-4 word description].ipynb
+ 2015-06-28-jw-initial-data-clean.html
+ 2015-06-28-jw-initial-data-clean.ipynb
+ 2015-06-28-jw-initial-data-clean.py
+ 2015-07-02-jw-coal-productivity-factors.html
+ 2015-07-02-jw-coal-productivity-factors.ipynb
+ 2015-07-02-jw-coal-productivity-factors.py
- deliver # (final analysis, code, presentations, etc)
+ Coal-mine-productivity.ipynb
+ Coal-mine-productivity.html
+ Coal-mine-productivity.py
- figures
+ 2015-07-16-jw-production-vs-hours-worked.png
- src # (modules and scripts)
+ init.py
+ load_coal_data.py
+ figures # (figures and plots)
+ production-vs-number-employees.png
+ production-vs-hours-worked.png
- data (backup-separate from version control)
+ coal_prod_cleaned.csv
Benefits
There are many benefits to this workflow and structure. The first and primary one is that they create a historical record of how the analysis progressed. It’s also easily searchable:
- by date (
ls 2015-06*.ipynb) - by author (
ls 2015*-jw-*.ipynb) - by topic (
ls *-coal-*.ipynb)
Second, during pull-requests, having the .py files lets a person quickly see which input text has changed, while having the .html files lets a person quickly see which outputs have changed. Having this be a painless post-save-hook makes this workflow effortless.
Finally, there are many smaller advantages of this approach that are too numerous to list here—please get in touch if you have questions, or suggestions for further improvements on the model!
Addendum
Jupyter 4.x is now out! If you use the incredibly useful Anaconda distribution, you can upgrade with conda update jupyter. More options covered here.
Further, config files are stored in ~/.jupyter by default. This directory is the JUPYTER_CONFIG_DIR environment variable. I’m going to first describe the above with the default config, and go into the complicated way of doing with Jupyter’s version of profiles.
The default config file is found at: ~/.jupyter/jupyter_notebook_config.py
If you don’t have this file, run: jupyter notebook --generate-config to create this file.
Add the following text to the top of the file:
c = get_config()
### If you want to auto-save .html and .py versions of your notebook:
# modified from: https://github.com/ipython/ipython/issues/8009
import os
from subprocess import check_call
def post_save(model, os_path, contents_manager):
"""post-save hook for converting notebooks to .py scripts"""
if model['type'] != 'notebook':
return # only do this for notebooks
d, fname = os.path.split(os_path)
check_call(['jupyter', 'nbconvert', '--to', 'script', fname], cwd=d)
check_call(['jupyter', 'nbconvert', '--to', 'html', fname], cwd=d)
c.FileContentsManager.post_save_hook = post_save
And you will have the same functionality above. Run jupyter notebook and you’re ready to go!
If you want to have this saving .html and .py files only when using a particular “profile,” it’s a bit trickier as Jupyter doesn’t use the notion of profiles anymore (as far as I can tell).
First create a new profile name via a bash command line:
export JUPYTER_CONFIG_DIR=~/.jupyter_profile2
jupyter notebook --generate-config
This will create a new directory and file at ~/.jupyter_profile2/jupyter_notebook_config.py. Then run jupyter notebook and work as usual. To switch back to your default profile you will have to set (either by hand, shell function, or your .bashrc) back to: export JUPYTER_CONFIG_DIR=~/.jupyter
Happy Data Sciencing!